Predicting Subglacial Lakes in Antarctica¶
Jake Bova | Programming for Geographic Information Systems | Fall 2024
We know: Subglacial lakes are present in Antarctica.
We want to know: Where are they?
Hypothesis: Subglacial lakes are more likely to be present in specific regions of Antarctica.
Using various geospatial raster datasets, I will predict the possible presence of subglacial lakes in Antarctica.
Primary Sources¶
- Wright A, Siegert M. A fourth inventory of Antarctic subglacial lakes. Antarctic Science. 2012
- Davies B. Antarctic Subglacial Lakes. AntarcticGlaciers.org. June 22, 2020
- Livingstone SJ. et al. Potential Subglacial Lake locations and meltwater drainage pathways beneath the Antarctic and Greenland Ice Sheets. The Cryosphere. 2013;7(6):1721-1740. doi:10.5194/tc-7-1721-2013
- Goeller S. et al. Assessing the Subglacial Lake coverage of Antarctica. Annals of Glaciology. 2016
Package Imports¶
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import geopandas as gpd
import raster_tools as rt
from sklearn.ensemble import RandomForestClassifier, IsolationForest
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report, confusion_matrix, roc_curve, auc
import seaborn as sns
from pathlib import Path
import time
import datetime
# supress all warnings
import warnings
warnings.filterwarnings("ignore")
Data¶
WEB RASTERS (IMPORTANT): https://umontana.maps.arcgis.com/apps/instant/imageryviewer/index.html?appid=88c564444ef744f1a16095c1dc358eb6
QUANTARCTICA: https://www.npolar.no/quantarctica/
Input Data
| Name | Type | Info | Grid Size | Source |
|---|---|---|---|---|
| known_lakes | Vector | GDF of known lakes | 1km | Wright, A., and Siegert, M. (2012) |
| lakes_raster | Vector/Raster | Rasterized known lake points | 1km | Wright, A., and Siegert, M. (2012) |
| dem | Raster | Digitel elevation model | 1km | BEDMAP2 (Fretwell et al., BAS/The Cryosphere, 2013) |
| slope | Raster | DEM Slope | 1km | BEDMAP2 (derived) |
| ice_thickness | Raster | Ice sheet thickness | 1km | BEDMAP2 (Fretwell et al., BAS/The Cryosphere, 2013) |
| bed_elevation | Raster | DEM of Antarctic bed | 1km | BEDMAP2 (Fretwell et al., BAS/The Cryosphere, 2013) |
| bed_slope | Raster | DEM Bed Slope | 1km | BEDMAP2 (derived, raster_tools) |
| bed_flow_dir | Raster | Bed flow direction | 1km | BEDMAP2 (derived, QGIS) |
| bed_flow_acc | Raster | Bed flow accumulation | 1km | BEDMAP2 (derived, QGIS) |
| hp_flow_acc | Raster | Hydraulic Flow Acc. Gradient (log) | 1km | BEDMAP2 (derived from bed_elevation and ice_thickness, Shreve equation implemented using raster_tools) |
| ice_bound_prox | Vector/Raster | Global Promity to ice boundary | 1km | MEaSUREs Antarctic boundaries (rasterized and calculated with raster_tools) |
| radarsat | Raster | Radar backscatter imagery | 100m | RAMP RADARSAT mosaic (Jezek et al., NSIDC, 1999/2013) (reprojected and warped with raster_tools to DEM 1km grid) |
| ice_velocity | Raster | X and Y velocity in m/yr | 450m | MEaSUREs Antarctic velocity map v2 (Rignot et al., NSIDC, 2017) (reprojected and warped with raster_tools to DEM 1km grid) |
| subglacial_water_flux | Raster | Modelled subglacial water flux beneath the grounded ice sheet | 1km | Le Brocq et al., Nature Geoscience, 2013, derived from BEDMAP2 |
| prox_to_known_lake | Raster | Global Proximity to known lake points | 1km | Wright, A., and Siegert, M. (2012), calculated with raster_tools |
| possible_lake_areas | Raster | Filtered raster from slope and ice_bound_prox | 1km | Jake Bova (2024) |
Shreve's Equation: $$\text{Hydraulic Potential} = \rho_w g h + \rho_i g H$$ Where:
- $\rho_w$ = density of water
- $g$ = acceleration due to gravity
- $h$ = bed elevation
- $\rho_i$ = density of ice
- $H$ = ice thickness
Original possible lake areas from Jake Bova (2024) are filtered by slope < ~0.1 and proximity to ice boundary < 200km to create possible_lake_areas.
This is a simple method but it is was good starting point for further analysis.
Data Processing¶
def load_lake_data(lake_path):
"""Load and preprocess lake data"""
lakes = gpd.read_file(lake_path)
# Clean LENGTH_M column
lakes['LENGTH_M'] = (lakes['LENGTH_M']
.str.replace('28100 + 4000', '28100')
.str.replace('<', '')
.str.replace(' ', ''))
lakes['LENGTH_M'] = pd.to_numeric(lakes['LENGTH_M'], errors='coerce')
lakes['SAT_DETECT'] = np.where(lakes['CLASS'] == 'G', 1, 0)
return lakes
def load_raster_data(config):
"""Load all raster data based on configuration dictionary"""
rasters = {}
for name, path in config['raster_paths'].items():
try:
rasters[name] = rt.Raster(path)
except Exception as e:
print(f"Error loading {name}: {e}")
# Handle derived rasters
if 'bed_slope' not in rasters and 'bed_elevation' in rasters:
rasters['bed_slope'] = rt.surface.slope(rasters['bed_elevation'])
if 'slope' not in rasters and 'dem' in rasters:
rasters['slope'] = rt.surface.slope(rasters['dem'])
# Special processing for subglacial water flux
if 'subglacial_water_flux' in rasters:
rasters['subglacial_water_flux'] = process_water_flux(rasters['subglacial_water_flux'])
return rasters
def process_water_flux(water_flux):
"""Process subglacial water flux data"""
water_flux = np.log10(water_flux + 1)
return water_flux.where(water_flux != -np.inf, other=np.nan)
def find_threshold_areas(slope, hp_flow_acc, ice_bound_prox):
"""Identify possible lake areas based on thresholds"""
condition1 = slope < 0.15 # 0.15 degrees angle
condition2 = (hp_flow_acc > 6) & (hp_flow_acc < 10)
condition3 = ice_bound_prox < 200000
combined_mask = condition1 & condition2 & condition3
return combined_mask.astype('uint8')
def reproject_rasters(rasters, template_raster, config):
"""Reproject rasters to match template"""
for name in config['needs_reprojection']:
if name in rasters:
rasters[name] = rt.warp.reproject(
rasters[name],
template_raster.crs,
resample_method='bilinear',
resolution=1000
)
return rasters
def clip_rasters(rasters, clip_geometry):
"""Clip all rasters to geometry"""
return {name: rt.clipping.clip(clip_geometry, raster)
for name, raster in rasters.items()}
def vectorize_rasters(rasters):
"""Convert rasters to vectors"""
vectors = {}
for name, raster in rasters.items():
vectors[name] = raster.to_vector().compute()
if 'band' in vectors[name].columns:
vectors[name].drop(columns=['band', 'row', 'col'], inplace=True)
# Add coordinates for specific layers
if name in ['radarsat', 'ice_velocity']:
vectors[name]['x_m'] = vectors[name].geometry.x
vectors[name]['y_m'] = vectors[name].geometry.y
# Apply offsets
if name == 'radarsat':
vectors[name]['x_m'] -= 50
vectors[name]['y_m'] += 50
elif name == 'ice_velocity':
vectors[name]['x_m'] -= 275
vectors[name]['y_m'] -= 725
return vectors
def merge_vectors(vectors, config):
"""Merge all vector data"""
merged = None
for step in config['merge_steps']:
left = vectors[step['left']]
right = vectors[step['right']]
if step.get('spatial', True):
merged = gpd.sjoin(
left if merged is None else merged,
right,
how=step.get('how', 'inner'),
op='intersects',
rsuffix=f"_{step['right']}"
)
else:
merged = merged.merge(
right,
on=step.get('on', ['x_m', 'y_m']),
how=step.get('how', 'inner')
)
if 'drop_columns' in step:
merged.drop(columns=step['drop_columns'], inplace=True)
if 'rename_columns' in step:
merged.rename(columns=step['rename_columns'], inplace=True)
# Post-processing
if 'known_lake' in merged.columns:
merged['possible_lake_area'] = np.where(
merged.known_lake == 1,
1,
merged.possible_lake_area
)
return merged
def get_data():
# Configuration
config = {
'raster_paths': {
'dem': 'data/Quantarctica3/Quantarctica3/TerrainModels/BEDMAP2/bedmap2_surface.tif',
'bed_elevation': 'data/Quantarctica3/Quantarctica3/TerrainModels/BEDMAP2/bedmap2_bed.tif',
'ice_thickness': 'data/Quantarctica3/Quantarctica3/TerrainModels/BEDMAP2/bedmap2_thickness.tif',
'bed_flow_dir': 'data/bed_flow_dir.tif',
'bed_flow_acc': 'data/bed_flow_acc.tif',
'hp_flow_acc': 'data/ant_flow_acc_log10.tif',
'radarsat': 'data/Quantarctica3/Quantarctica3/SatelliteImagery/RADARSAT/RADARSAT_Mosaic.jp2',
'ice_velocity': 'input_data/icevel_filled.tif',
'subglacial_water_flux': 'data/Quantarctica3/Quantarctica3/Glaciology/Subglacial Water Flux/SubglacialWaterFlux_Modelled_1km.tif'
},
'needs_reprojection': ['radarsat', 'ice_velocity'],
'merge_steps': [
{
'left': 'slope',
'right': 'dem',
'rename_columns': {'value_slope': 'slope', 'value_dem': 'dem'},
'drop_columns': ['band_slope', 'row_slope', 'col_slope', 'index_dem', 'band_dem', 'row_dem', 'col_dem']
},
{
'left': 'merged',
'right': 'ice_thickness',
'rename_columns': {'value': 'ice_thickness'}
},
{
'left': 'merged',
'right': 'bed_elevation',
'rename_columns': {'value': 'bed_elevation'},
'drop_columns': ['index_ice_thickness', 'band_left', 'row_left', 'col_left',
'index_bed_elevation', 'band_bed_elevation', 'row_bed_elevation', 'col_bed_elevation']
},
{
'left': 'merged',
'right': 'bed_slope',
'rename_columns': {'value': 'bed_slope'},
'drop_columns': ['index_bed_slope', 'band', 'row', 'col']
},
{
'left': 'merged',
'right': 'bed_flow_dir',
'rename_columns': {'value': 'bed_flow_dir'},
'drop_columns': ['index_bed_flow_dir', 'band', 'row', 'col']
},
{
'left': 'merged',
'right': 'bed_flow_acc',
'rename_columns': {'value': 'bed_flow_acc'},
'drop_columns': ['index_bed_flow_acc', 'band', 'row', 'col']
},
{
'left': 'merged',
'right': 'hp_flow_acc',
'rename_columns': {'value': 'hp_flow_acc_log'},
'drop_columns': ['index_hp_flow_acc', 'band', 'row', 'col']
},
{
'left': 'merged',
'right': 'prox_to_known_lake',
'rename_columns': {'value': 'prox_to_known_lake_m'},
'drop_columns': ['index_prox_to_known_lake', 'band', 'row', 'col']
},
{
'left': 'merged',
'right': 'ice_bound_prox',
'rename_columns': {'value': 'ice_bound_prox_m'},
'drop_columns': ['index_ice_bound_prox', 'band', 'row', 'col']
},
{
'left': 'merged',
'right': 'lakes',
'how': 'left',
'rename_columns': {'value': 'known_lake'},
'drop_columns': ['index_lakes', 'band', 'row', 'col']
},
{
'left': 'merged',
'right': 'possible_lake_areas',
'rename_columns': {'value': 'possible_lake_area'},
'drop_columns': ['index_possible_lake_areas', 'band', 'row', 'col']
},
{
'left': 'merged',
'right': 'subglacial_water_flux',
'rename_columns': {'value': 'subglacial_water_flux'},
'drop_columns': ['index_subglacial_water_flux', 'band', 'row', 'col']
},
{
'left': 'merged',
'right': 'radarsat',
'spatial': False,
'rename_columns': {'value': 'radarsat', 'geometry_x': 'geometry'},
'drop_columns': ['geometry_y']
},
{
'left': 'merged',
'right': 'ice_velocity',
'spatial': False,
'rename_columns': {'value': 'ice_vel', 'geometry_x': 'geometry'},
'drop_columns': ['geometry_y']
}
]
}
# Load and process data
lakes = load_lake_data('data/Quantarctica3/Quantarctica3/Glaciology/Subglacial Lakes/SubglacialLakes_WrightSiegert.shp')
rasters = load_raster_data(config)
# Load world boundary for clipping
world = gpd.read_file('data/Quantarctica3/Quantarctica3/Miscellaneous/SimpleBasemap/ADD_DerivedLowresBasemap.shp')
world = world.to_crs(epsg=3031)
world_land = world[world['Category'] == 'Land']
# Process rasters
rasters = reproject_rasters(rasters, rasters['dem'], config)
# Calculate possible lake areas
rasters['possible_lake_areas'] = find_threshold_areas(
rasters['slope'],
rasters['hp_flow_acc'],
rasters['ice_bound_prox']
)
# Clip and vectorize
clipped_rasters = clip_rasters(rasters, world_land)
vectors = vectorize_rasters(clipped_rasters)
# Merge data
merged = merge_vectors(vectors, config)
# Final processing
merged = gpd.GeoDataFrame(merged, geometry='geometry')
merged.set_crs(epsg=3031, inplace=True)
# Clean up
merged = merged.dropna(subset=['slope'])
merged['subglacial_water_flux'] = merged['subglacial_water_flux'].fillna(
merged['subglacial_water_flux'].mean()
)
merged = merged.dropna(subset=['subglacial_water_flux'])
return merged
# merged = get_data() # Load data
merged = pd.read_csv('input_data/merged_data.csv')
merged = gpd.GeoDataFrame(merged, geometry=gpd.points_from_xy(merged.x_m, merged.y_m))
merged.set_crs(epsg=3031, inplace=True)
merged.drop(columns=['Unnamed: 0'], inplace=True)
df = merged.drop(columns=['geometry', 'prox_to_known_lake_m', 'possible_lake_area', 'x_m', 'y_m']) # Drop geometry and other columns for training (x_m and y_m were used for merging)
df_known_lakes = df[df['known_lake'] == 1]
After preparing the data we get a geodataframe of every raster point covered by BEDMAP2.
I decided to not use proximity to known lakes as a feature because it is basically the same as the known_lakes raster (with 0 distance being a known lake).
df
| slope | dem | ice_thickness | bed_elevation | bed_slope | bed_flow_dir | bed_flow_acc | hp_flow_acc_log | ice_bound_prox_m | known_lake | subglacial_water_flux | radarsat | ice_vel | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.707877 | 51 | 168 | -117 | 0.780114 | 8 | 2.0 | 5.560527 | 1000.0000 | 0 | 0.0 | 218 | 9.844176 |
| 1 | 0.810800 | 56 | 164 | -108 | 0.683401 | 8 | 3.0 | 5.981568 | 1000.0000 | 0 | 0.0 | 220 | 3.164224 |
| 2 | 0.907321 | 60 | 162 | -102 | 0.567633 | 8 | 3.0 | 6.260069 | 1414.2136 | 0 | 0.0 | 223 | 2.593423 |
| 3 | 0.999757 | 62 | 160 | -98 | 0.489942 | 8 | 0.0 | 6.478846 | 1414.2136 | 0 | 0.0 | 224 | 3.497082 |
| 4 | 1.088866 | 63 | 160 | -97 | 0.452274 | 4 | 0.0 | 6.663968 | 1000.0000 | 0 | 0.0 | 224 | 3.351763 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 11859923 | 3.375342 | 158 | 344 | -186 | 3.351585 | 4 | 3.0 | 6.926358 | 1000.0000 | 0 | 0.0 | 217 | 8.147156 |
| 11859924 | 3.489445 | 178 | 339 | -161 | 3.467022 | 4 | 2.0 | 6.784882 | 1000.0000 | 0 | 0.0 | 206 | 5.284122 |
| 11859925 | 3.480071 | 192 | 334 | -142 | 3.459608 | 4 | 1.0 | 6.977334 | 1414.2136 | 0 | 0.0 | 208 | 3.320855 |
| 11859926 | 3.348109 | 198 | 327 | -129 | 3.278850 | 4 | 0.0 | 6.818604 | 1000.0000 | 0 | 0.0 | 214 | 2.504690 |
| 11859927 | 3.147901 | 192 | 321 | -129 | 3.007938 | 4 | 0.0 | 6.210092 | 1000.0000 | 0 | 0.0 | 222 | 2.143291 |
11859928 rows × 13 columns
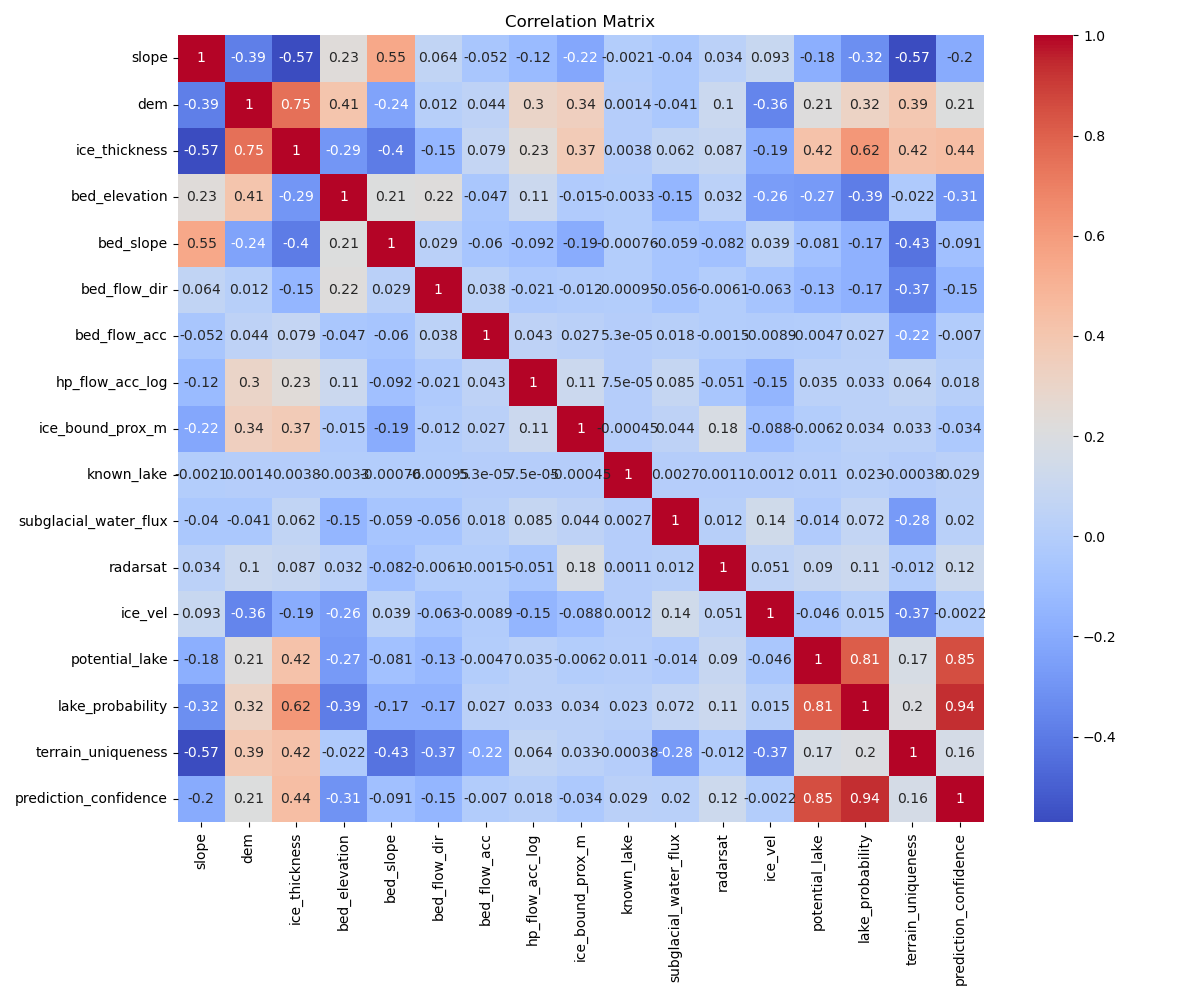
Many features correlate with each other, as can be seen from the correlation matrix below. This is expected as geospacial data is often highly correlated, and many of the features are derived from the same data.
df_no_lakes = df.drop(columns=['known_lake'])
corr = df_no_lakes.corr()
plt.figure(figsize=(12, 10))
sns.heatmap(corr, annot=True, cmap='coolwarm', center=0)
plt.show()
We can also inspect the distribution of data in known lake areas vs the rest of the data.
fig, axes = plt.subplots(4, 3, figsize=(15, 15))
cols = df.columns
cols = cols.drop('known_lake')
for i, col in enumerate(cols):
ax = axes[i // 3, i % 3]
# Calculate statistics
stats_all = {
'mean': df[col].mean(),
'median': df[col].median(),
'min': df[col].min(),
'max': df[col].max()
}
stats_known = {
'mean': df_known_lakes[col].mean(),
'median': df_known_lakes[col].median(),
'min': df_known_lakes[col].min(),
'max': df_known_lakes[col].max()
}
# Plot histograms
ax.hist(df[col], bins=50, alpha=0.5, label='All', log=True, color='orange')
ax.hist(df_known_lakes[col], bins=50, alpha=0.5, label='Known Lakes', log=True, color='blue')
# Add title and legend
ax.set_title(col)
ax.legend(loc='upper right')
# Create and add text blocks separately with different colors
all_text = (
f"All Data (orange):\n"
f" Mean: {stats_all['mean']:.2f}\n"
f" Median: {stats_all['median']:.2f}\n"
f" Range: [{stats_all['min']:.2f}, {stats_all['max']:.2f}]"
)
known_text = (
f"\n\nKnown Lakes (blue):\n"
f" Mean: {stats_known['mean']:.2f}\n"
f" Median: {stats_known['median']:.2f}\n"
f" Range: [{stats_known['min']:.2f}, {stats_known['max']:.2f}]"
)
# Add text blocks with separate colors
ax.text(0.05, 0.95, all_text,
transform=ax.transAxes,
verticalalignment='top',
fontsize=8,
family='monospace',
color='orange',
bbox=dict(facecolor='white', alpha=0.8))
ax.text(0.05, 0.65, known_text,
transform=ax.transAxes,
verticalalignment='top',
fontsize=8,
family='monospace',
color='blue',
bbox=dict(facecolor='white', alpha=0.8))
plt.tight_layout()
plt.show()
As you can see, the data for known lake locations follows a similar distribution to the rest of the data, but is skewed towards extreme values across almost all features.
Model Creation¶
I decided to use a Random Forest model, as it is a good starting point for geospatial data and I have had success with it in the past.
Simple initial model¶
# simple random forest model to predict known_lake
# we will have to oversample the known_lake = 1 class
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report, confusion_matrix
# first, drop n percent of the data that has known_lake = 0
df_sample = df.sample(frac=0.2)
# add back the known_lakes 10 times
for i in range(10):
df_sample = pd.concat([df_sample, df_known_lakes])
X = df_sample.drop(columns=['known_lake'])
y = df_sample['known_lake']
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3)
# oversample the known_lake = 1 class
from imblearn.over_sampling import SMOTE
# sm = SMOTE(random_state=42)
# X_train, y_train = sm.fit_resample(X_train, y_train)
clf = RandomForestClassifier(n_estimators=100)
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
print(confusion_matrix(y_test, y_pred))
print(classification_report(y_test, y_pred))
# get column names and then list the feature importances
feature_importances = clf.feature_importances_
feature_names = X.columns
feature_importances_df = pd.DataFrame({'feature': feature_names, 'importance': feature_importances})
feature_importances_df = feature_importances_df.sort_values('importance', ascending=False)
feature_importances_df
Enhanced model¶
def prepare_data(df, sample_frac=0.05, known_lakes_multiplier=10):
"""
Prepare training data with balanced representation of known lakes and non-lake areas
Args:
df: Input dataframe containing lake and terrain features
sample_frac: Fraction of non-lake data to sample (reduces class imbalance)
known_lakes_multiplier: Number of times to duplicate known lake data (further balances classes)
"""
# Take a random sample of the full dataset to reduce computational load and class imbalance
df_sample = df.sample(frac=sample_frac, random_state=42)
# Extract all known lake locations
df_known_lakes = df[df['known_lake'] == 1]
# Since we are sampling such a small fraction of the data, we need to balance the classes
# I was using SMOTE before, but just duplicating the known lakes is simpler and works well
# It does cause some overfitting, but that is kind of the point here (we want to find lakes)
# Duplicate known lakes multiple times to balance the dataset
# This helps prevent the model from being biased towards the majority class (non-lakes)
for i in range(known_lakes_multiplier):
df_sample = pd.concat([df_sample, df_known_lakes])
# Remove any rows with missing values to ensure clean training data
df_sample.dropna(inplace=True)
return df_sample
def train_anomaly_detector(X_train, contamination=0.01):
"""
Train an isolation forest to detect unusual terrain features
The idea here is to identify areas that are unique compared to the rest of the dataset.
Imagine this as a way to find "hidden lakes" that are not easily classified by the main model.
The basic idea is that if a location is very different from the rest of the terrain, it might be a lake (naive assumption).
We basically look at every feature of a data point and see how different it is from the rest of the dataset.
Args:
X_train: Training data with terrain features
contamination: Expected proportion of anomalies in the dataset
"""
# Initialize isolation forest with specified contamination rate
# Contamination represents expected proportion of outliers in the dataset
iso_forest = IsolationForest(contamination=contamination, random_state=42)
# Fit the model to learn what "normal" terrain looks like
iso_forest.fit(X_train)
return iso_forest
def create_lake_prediction_model(df, features_to_exclude=None, data_frac=0.05):
"""
Create and train both the main classifier and anomaly detector
Args:
df: Input dataframe with all features
features_to_exclude: List of columns to exclude from training
data_frac: Fraction of data to use for training
"""
# Get balanced dataset for training
df_sample = prepare_data(df, sample_frac=data_frac)
# Separate features and target variable
X = df_sample.drop(columns=features_to_exclude if features_to_exclude else [])
y = df_sample['known_lake']
# Standardize features to ensure all are on same scale
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
X_scaled = pd.DataFrame(X_scaled, columns=X.columns)
# Split data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(X_scaled, y, test_size=0.3, random_state=42)
# Initialize and train Random Forest classifier with parameters tuned for imbalanced data
clf = RandomForestClassifier(n_estimators=200, # Number of trees
max_depth=10, # Maximum depth of trees (prevents overfitting)
min_samples_split=5, # Minimum samples needed to split a node
class_weight='balanced', # Adjusts weights inversely proportional to class frequencies
random_state=42)
clf.fit(X_train, y_train)
# Train anomaly detector on same training data
iso_forest = train_anomaly_detector(X_train)
return clf, iso_forest, scaler, X_test, y_test
def predict_potential_lakes(df, clf, iso_forest, scaler, features_to_exclude=None,
similarity_threshold=0.3, anomaly_threshold=-0.5):
"""
Combine classifier and anomaly detection predictions to identify potential lakes
Args:
similarity_threshold: Minimum probability threshold for lake classification
anomaly_threshold: Minimum score for terrain uniqueness
"""
# Clean and prepare prediction data
df.dropna(inplace=True)
X_pred = df.drop(columns=features_to_exclude if features_to_exclude else [])
X_pred_scaled = scaler.transform(X_pred)
# Get probability predictions from random forest (how likely is this a lake?)
lake_probabilities = clf.predict_proba(X_pred_scaled)[:, 1]
# Get anomaly scores (how unique is this terrain?)
anomaly_scores = iso_forest.score_samples(X_pred_scaled)
# Combine predictions: location must have both high lake probability and unique terrain
potential_lakes = (lake_probabilities > similarity_threshold) & (anomaly_scores < anomaly_threshold) # swapped from > to <, because anomaly score is negative, meaning lower is more unique
return potential_lakes, lake_probabilities, anomaly_scores
def analyze_predictions(df, potential_lakes, lake_probabilities, anomaly_scores, iteration=None):
"""
Combine predictions and calculate confidence scores
Args:
iteration: Used for iterative confidence weighting (optional)
"""
# Copy input data to avoid modifications
df_results = df.copy()
# Add prediction results as new columns
df_results['potential_lake'] = potential_lakes
df_results['lake_probability'] = lake_probabilities
df_results['terrain_uniqueness'] = anomaly_scores
# Calculate confidence score as weighted combination of lake probability and terrain uniqueness
df_results['prediction_confidence'] = (
(df_results['lake_probability'] * 0.7) + # Higher weight to classifier prediction
(df_results['terrain_uniqueness'] * 0.3) # Lower weight to terrain uniqueness (also, is negative, so this is a penalty)
).clip(0, 1) # Ensure confidence is between 0 and 1
return df_results
def evaluate_model(clf, X_test, y_test, feature_names=None, fname=False, iteration=None):
"""
Comprehensive model evaluation with metrics and visualizations
Args:
fname: Whether to save plots to file
iteration: Used for saving multiple versions of plots
"""
# Get model predictions on test set
y_pred = clf.predict(X_test)
y_pred_proba = clf.predict_proba(X_test)[:, 1]
# Print standard classification metrics
print("\nClassification Report:")
print(classification_report(y_test, y_pred))
# Create and plot confusion matrix
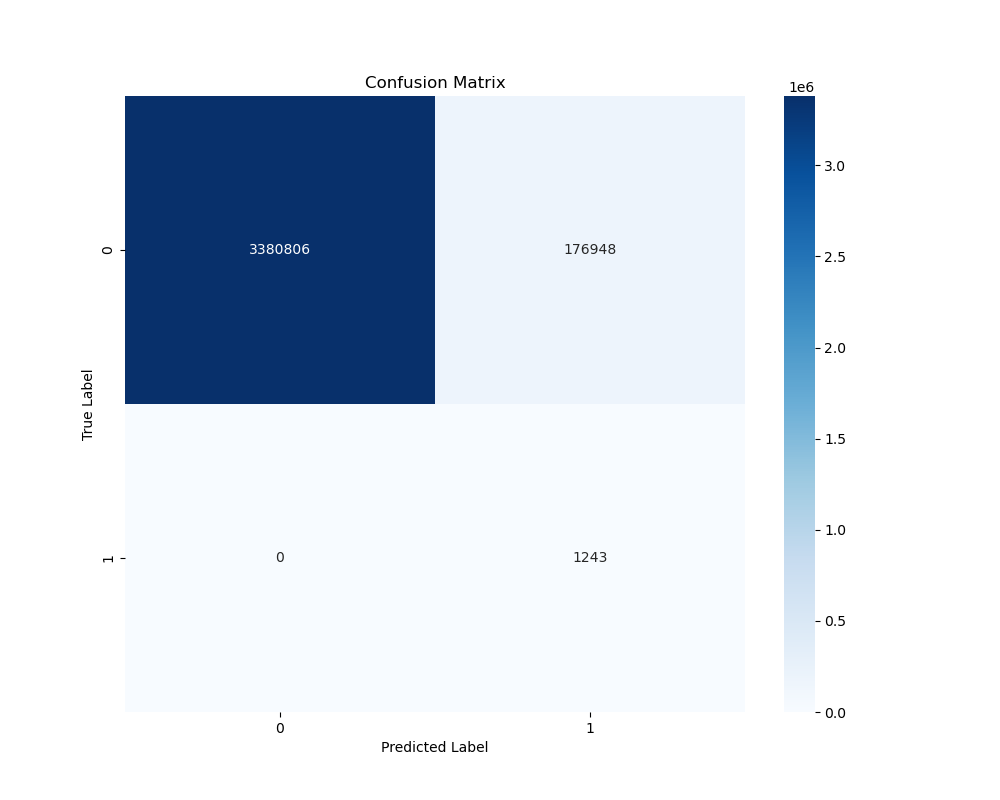
plt.figure(figsize=(10, 8))
cm = confusion_matrix(y_test, y_pred)
sns.heatmap(cm, annot=True, fmt='d', cmap='Blues')
plt.title('Confusion Matrix')
plt.ylabel('True Label')
plt.xlabel('Predicted Label')
if fname:
plt.savefig(f'results/confusion_matrix_{iteration}.png')
else:
plt.show()
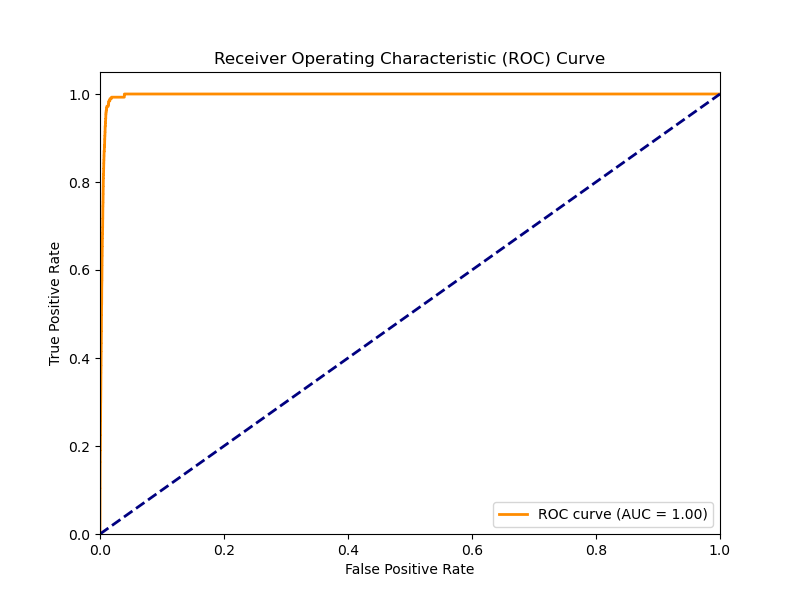
# Plot ROC curve and calculate AUC
plt.figure(figsize=(8, 6))
fpr, tpr, _ = roc_curve(y_test, y_pred_proba)
roc_auc = auc(fpr, tpr)
plt.plot(fpr, tpr, color='darkorange', lw=2,
label=f'ROC curve (AUC = {roc_auc:.2f})')
plt.plot([0, 1], [0, 1], color='navy', lw=2, linestyle='--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver Operating Characteristic (ROC) Curve')
plt.legend(loc="lower right")
if fname:
plt.savefig(f'results/roc_curve_{iteration}.png')
else:
plt.show()
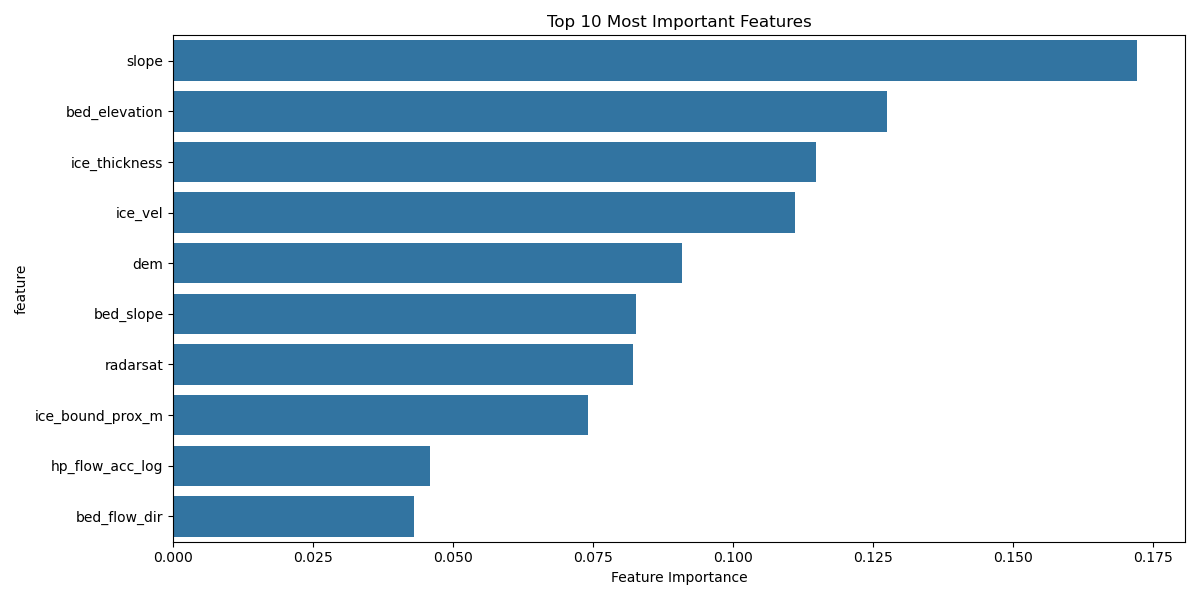
# Create feature importance visualization
if feature_names is not None:
importance = pd.DataFrame({
'feature': feature_names,
'importance': clf.feature_importances_
}).sort_values('importance', ascending=False)
plt.figure(figsize=(12, 6))
sns.barplot(data=importance.head(10), x='importance', y='feature')
plt.title('Top 10 Most Important Features')
plt.xlabel('Feature Importance')
plt.tight_layout()
if fname:
plt.savefig(f'results/feature_importance_{iteration}.png')
else:
plt.show()
return importance
def visualize_predictions_dual(results_gdf, min_probability=0.3, fname=False, iteration=None):
"""
Create two-panel visualization comparing lake probabilities and prediction confidence
Args:
results_gdf: GeoDataFrame with prediction results
min_probability: Minimum probability threshold for displaying predictions
"""
# Create two-panel figure
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20, 8))
# Filter to show only predictions above minimum probability threshold
potential_lakes = results_gdf[results_gdf['lake_probability'] >= min_probability].copy()
# Panel 1: Lake Probability
# Plot low-probability points as background
ax1.scatter(results_gdf[results_gdf['lake_probability'] < min_probability].geometry.x,
results_gdf[results_gdf['lake_probability'] < min_probability].geometry.y,
c='gray', alpha=0.1, s=1)
# Plot predicted lakes colored by probability
scatter1 = ax1.scatter(potential_lakes.geometry.x,
potential_lakes.geometry.y,
c=potential_lakes['lake_probability'],
cmap='viridis',
s=30,
alpha=0.7)
# Highlight known lakes with red circles
ax1.scatter(results_gdf[results_gdf['known_lake'] == 1].geometry.x,
results_gdf[results_gdf['known_lake'] == 1].geometry.y,
s=25, marker='o', label='Known lakes', alpha=0.3, facecolors='none', edgecolors='red')
# Add colorbar and labels
plt.colorbar(scatter1, ax=ax1, label='Lake Probability')
ax1.set_title('Lake Probability Distribution')
ax1.set_xlabel('X Coordinate (m)')
ax1.set_ylabel('Y Coordinate (m)')
ax1.legend()
ax1.grid(True, alpha=0.3)
# Panel 2: Prediction Confidence
# Repeat similar plotting for confidence scores
ax2.scatter(results_gdf[results_gdf['lake_probability'] < min_probability].geometry.x,
results_gdf[results_gdf['lake_probability'] < min_probability].geometry.y,
c='gray', alpha=0.1, s=1)
scatter2 = ax2.scatter(potential_lakes.geometry.x,
potential_lakes.geometry.y,
c=potential_lakes['prediction_confidence'],
cmap='viridis',
s=30,
alpha=0.7)
ax2.scatter(results_gdf[results_gdf['known_lake'] == 1].geometry.x,
results_gdf[results_gdf['known_lake'] == 1].geometry.y,
s=25, marker='o', label='Known lakes', alpha=0.3, facecolors='none', edgecolors='red')
plt.colorbar(scatter2, ax=ax2, label='Prediction Confidence')
ax2.set_title('Prediction Confidence Distribution')
ax2.set_xlabel('X Coordinate (m)')
ax2.set_ylabel('Y Coordinate (m)')
ax2.legend()
ax2.grid(True, alpha=0.3)
plt.tight_layout()
if fname:
plt.savefig(f'results/spatial_predictions_{iteration}.png')
else:
plt.show()
Model Usage¶
for i in range(1): # iterative setup to test different parameters
print(f'## Iteration {i} ##')
print('## Creating Lake Prediction Model ##')
clf, iso_forest, scaler, X_test, y_test = create_lake_prediction_model(df, features_to_exclude=['known_lake'], data_frac=0.1)
print('## Model Created ##')
print('## Predicting Potential Lakes ##')
potential_lakes, probs, scores = predict_potential_lakes(df, clf, iso_forest, scaler,
features_to_exclude=['known_lake'], similarity_threshold=0.3, anomaly_threshold=-0.4)
print('## Analyzing Predictions ##')
results = analyze_predictions(df, potential_lakes, probs, scores, iteration=i)
# Evaluate model
feature_names = df.drop(columns=['known_lake']).columns
print('## Evaluating Model ##')
importance_df = evaluate_model(clf, X_test, y_test, feature_names, fname=False, iteration=i)
# Merge with geometry and create GeoDataFrame
print('## Merging with Geometry ##')
results = results.merge(merged['geometry'], left_index=True, right_index=True)
results_gdf = gpd.GeoDataFrame(results, geometry='geometry')
# visualize_predictions_dual(results_gdf, min_probability=0.3, fname=False, iteration=i)
 |
 |
 |
 |
results_known_lakes = results[results['known_lake'] == 1]
results_pred_lakes = results[results['potential_lake'] == 1]
fig, axes = plt.subplots(1, 3, figsize=(15, 5))
cols = ['lake_probability', 'terrain_uniqueness', 'prediction_confidence']
for i, col in enumerate(cols):
ax = axes[i]
# Calculate statistics
stats_all = {
'mean': results_gdf[col].mean(),
'median': results_gdf[col].median(),
'min': results_gdf[col].min(),
'max': results_gdf[col].max()
}
stats_pred = {
'mean': results_pred_lakes[col].mean(),
'median': results_pred_lakes[col].median(),
'min': results_pred_lakes[col].min(),
'max': results_pred_lakes[col].max()
}
stats_known = {
'mean': results_known_lakes[col].mean(),
'median': results_known_lakes[col].median(),
'min': results_known_lakes[col].min(),
'max': results_known_lakes[col].max()
}
# Plot histograms
ax.hist(results_gdf[col], bins=50, alpha=0.5, label='All', log=True, color='orange')
ax.hist(results_pred_lakes[col], bins=50, alpha=0.5, label='Predicted Lakes', log=True, color='grey')
ax.hist(results_known_lakes[col], bins=50, alpha=0.5, label='Known Lakes', log=True, color='blue')
# Add title and legend
ax.set_title(col)
ax.legend(loc='upper right')
# Create and add text blocks separately with different colors
all_text = (
f"All Data (orange):\n"
f" Mean: {stats_all['mean']:.2f}\n"
f" Median: {stats_all['median']:.2f}\n"
f" Range: [{stats_all['min']:.2f}, {stats_all['max']:.2f}]"
)
pred_text = (
f"\n\nPredicted Lakes (grey):\n"
f" Mean: {stats_pred['mean']:.2f}\n"
f" Median: {stats_pred['median']:.2f}\n"
f" Range: [{stats_pred['min']:.2f}, {stats_pred['max']:.2f}]"
)
known_text = (
f"\n\nKnown Lakes (blue):\n"
f" Mean: {stats_known['mean']:.2f}\n"
f" Median: {stats_known['median']:.2f}\n"
f" Range: [{stats_known['min']:.2f}, {stats_known['max']:.2f}]"
)
# Add text blocks with separate colors
ax.text(0.05, 0.95, all_text,
transform=ax.transAxes,
verticalalignment='top',
fontsize=8,
family='monospace',
color='orange',
bbox=dict(facecolor='white', alpha=0.8))
ax.text(0.05, 0.65, known_text,
transform=ax.transAxes,
verticalalignment='top',
fontsize=8,
family='monospace',
color='blue',
bbox=dict(facecolor='white', alpha=0.8))
plt.tight_layout()
plt.show()
To determine the binary classification of possible lake areas, I used a lake probability threshold of 0.4 and a terrain uniqueness threshold of -0.4.
These values were determined using trial and error as well as researched information on subglacial lakes.
My findings compared to those of Goeller S. et al. (2016) show that my model is more conservative in predicting subglacial lakes. This is likely due to the terrain uniqueness threshold I used.
My final prediction map.